Redesign 2: Rats of New York
Due by 11:59 PM on Monday, October 2, 2017
New York City is full of urban wildlife, and rats are one of the city’s most infamous animal mascots. Rats in NYC are plentiful, but they also deliver food, so they’re useful too.
NYC keeps incredibly detailed data regarding animal sightings, including rats, and it makes this data publicly available.
For this second redesign assignment, you will use R and ggplot to tell an interesting story hidden in the data. You can recreate one of these ugly, less-than-helpful graphs, or create a new story by looking at other variables in the data:
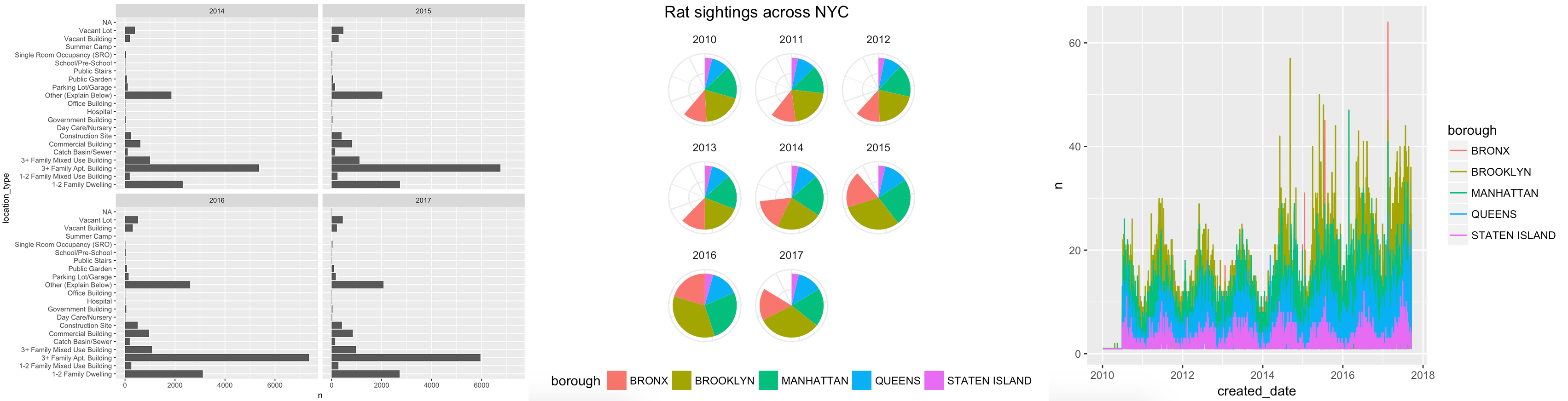
Here’s what you need to do:
- Download New York City’s database of rat sightings since 2010:

NYC Rat Sightings. Place this in a folder nameddatain an RStudio project.As always, you’ll probably need to right click on this link and choose “Save link as…”, since your browser will want to display it as text. The data was originally uploaded by the City of New York to Kaggle, and is provided with a public domain license.
- Summarize the data somehow. The raw data has more than 100,000 rows, which means you’ll need to aggregate the data. Consider looking at the number of sightings per borough, per year, per dwelling type, etc., or a combination of these, like the change in the number sightings across the 5 boroughs between 2010 and 2016.
- Create an appropriate visualization based on the data you summarized.
- Write a memo (no word limit) explaining your process. I’m specifically looking for a discussion of the following:
- What was wrong with the original graphic (if you’re fixing one of the original figures)?
- What story are you telling with your new graphic?
- How did you apply the principles of CRAP?
- How did you apply Alberto Cairo’s five qualities of great visualizations?
- E-mail me the following outputs:
- A PDF of your memo with your final code and graphic embedded in it.You can approach this in a couple different ways—you can write the memo and then include the full figure and code at the end, similar to this blog post, or you can write the memo in an incremental way, describing the different steps of creating the figure, ultimately arriving at a clean final figure, like this blog post.
This means you’ll need to do all your coding in an R Markdown file and embed your code in chunks. - A standalone PNG version of your graphicUse
ggsave(plot_name, filename = "blah.png", width = XX, height = XX)
- A standalone PDF version of your graphicUse
ggsave(plot_name, filename = "blah.pdf", width = XX, height = XX)
- A PDF of your memo with your final code and graphic embedded in it.You can approach this in a couple different ways—you can write the memo and then include the full figure and code at the end, similar to this blog post, or you can write the memo in an incremental way, describing the different steps of creating the figure, ultimately arriving at a clean final figure, like this blog post.
You will be graded based on how you use R and ggplot, how well you apply the principles of CRAP, The Truthful Art, and Effective Data Visualization, and how appropriate the graph is for the data and the story you’re telling. I will use this rubric to grade the final product. 
For this assignment, I am less concerned with detailed graphic design principles—select appropriate colors, change fonts if you’re brave, and choose a nice ggplot theme and perhaps make a few adjustments like moving the legend around (theme(legend.position = "bottom")) or other things you find with ggThemeAssist.
The assignment is due by midnight on Monday, October 2.
This assignment is more code-intensive than the first redesign assignment, but you have enough R skills to do this (really really). Help abounds—work in groups,Though your assignments have to be turned in individually…
consult with your classmates, meet with me, and search Google. You can do this, and you’ll hopefully feel like a dataviz wizard/witch when you’re done.
I’ve provided some starter code below. A couple comments about it:
- By default,
read_csv()treats cells that are empty or “NA” as missing values. This rat dataset uses “N/A” to mark missing values, so we need to add that as a possible marker of missingness (hencena = c("", "NA", "N/A")) - To make life easier, I’ve renamed some of the key variables you might work with. You can rename others if you want.
- I’ve also created a few date-related variables (
sighting_year,sighting_month,sighting_day, andsighting_weekday). You don’t have to use them, but they’re there if you need them.The functions that create these, likeyear()andwday()are part of thelubridatelibrary.
- The date/time variables are formatted like
04/03/2017 12:00:00 AM, which R is not able to automatically parse as a date when reading the CSV file. You can use themdy_hms()function in thelubridatelibrary to parse dates that are structured as “month-day-year-hour-minute.”There are also a bunch of other iterations of this function, likeymd(),dmy(), etc., for other date formats.
- There’s one row with an unspecified borough, so I filter that out.
library(tidyverse)
library(lubridate)
rats_raw <- read_csv("data/Rat_Sightings.csv", na = c("", "NA", "N/A"))
# If you get an arror that says "All formats failed to parse. No formats
# found", it's because the mdy_hms function couldn't parse the date. The date
# variable *should* be in this format: "04/03/2017 12:00:00 AM", but in some
# rare instances, it might load without the seconds as "04/03/2017 12:00 AM".
# If there are no seconds, use mdy_hm() instead of mdy_hms().
rats_clean <- rats_raw %>%
rename(created_date = `Created Date`,
location_type = `Location Type`,
borough = Borough) %>%
mutate(created_date = mdy_hm(created_date)) %>%
mutate(sighting_year = year(created_date),
sighting_month = month(created_date),
sighting_day = day(created_date),
sighting_weekday = wday(created_date, label = TRUE, abbr = FALSE)) %>%
filter(borough != "Unspecified")You’ll summarize the data with functions from dplyr, including stuff like count(), arrange(), filter(), group_by(), summarize(), and mutate(). Here are some examples of ways to summarize the data:
# See the count of rat sightings by weekday
rats_clean %>%
count(sighting_weekday)
# Assign a table to an object to use it in a plot
rats_by_weekday <- rats_clean %>%
count(sighting_weekday, sighting_year)
# geom_bar() will try to calculate the count for you by default. Here we
# already have the count, so we have to specify that the statistic we care
# about in the bar chart is "identity", or the actual variable that's there
# already.
#
# coord_flip() flips the plot so that the weekdays show up on the y-axis. This
# messes with the ordering, though, and puts Saturday at the top. You can
# reverse the levels of the weekday category with the fct_rev function in the
# forcats package, which provides a bunch of functions for working with
# factors.
ggplot(rats_by_weekday, aes(x = forcats::fct_rev(sighting_weekday), y = n)) +
geom_bar(stat = "identity") +
coord_flip() +
facet_wrap(~ sighting_year)
# See the count of rat sightings by weekday and borough
rats_clean %>%
count(sighting_weekday, borough, sighting_year)
# An alternative to count() is to specify the groups with group_by() and then
# be explicit about how you're summarizing the groups, such as calculating the
# mean, standard deviation, or number of observations (we do that here with
# `n()`).
rats_clean %>%
group_by(sighting_weekday, borough) %>%
summarize(n = n())